*A guide to scikit-learn compatible nearest neighbors classification using the recently introduced word mover’s distance (WMD). * Joint post with the awesome Matt Kusner!
Source of this Jupyter notebook.
In document classification and other natural language processing applications, having a good measure of the similarity of two texts can be a valuable building block. Ideally, such a measure would capture semantic information. Cosine similarity on bag-of-words vectors is known to do well in practice, but it inherently cannot capture when documents say the same thing in completely different words.
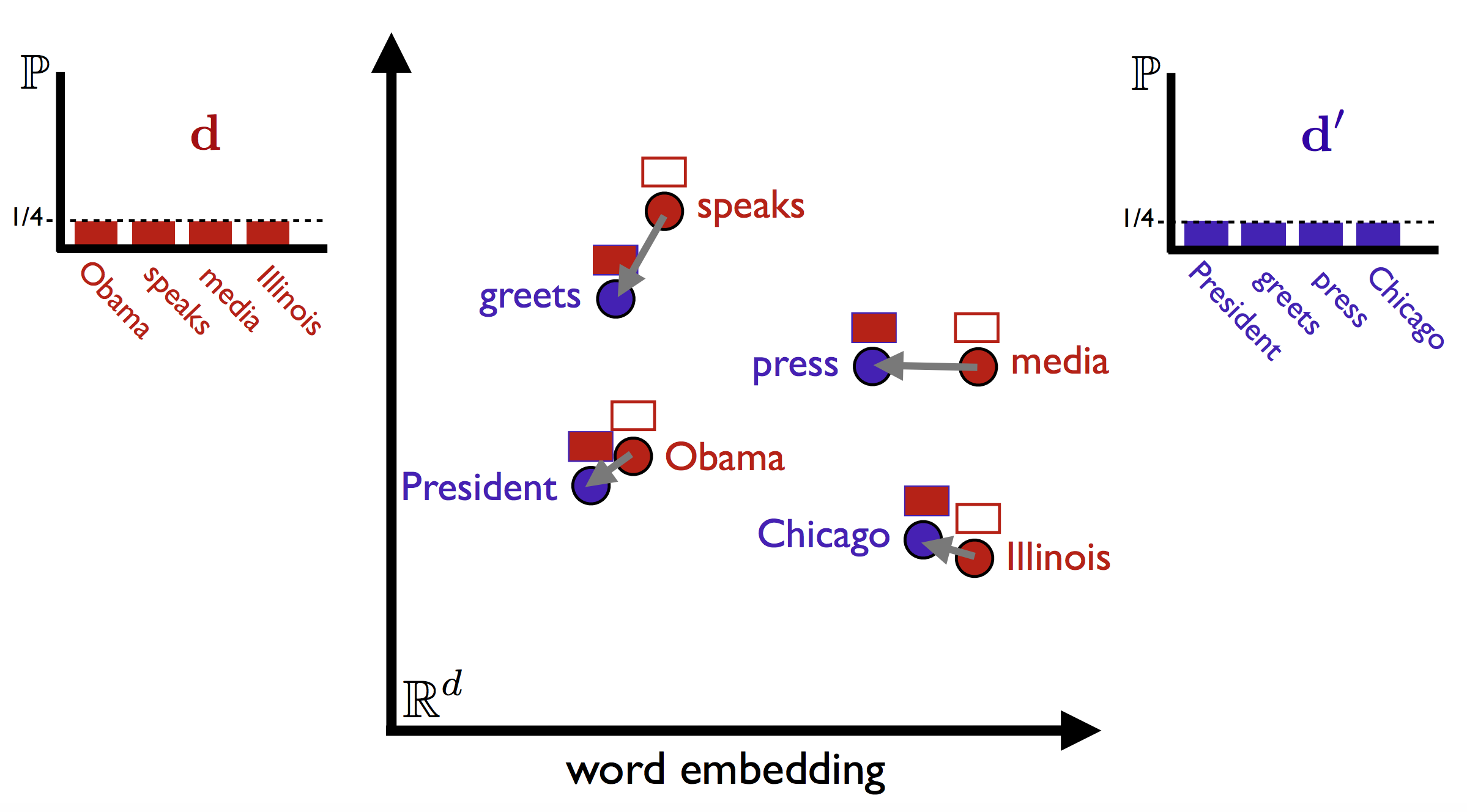
Take, for example, two headlines:
- Obama speaks to the media in Illinois
- The President greets the press in Chicago
These have no content words in common, so according to most bag of words—based metrics, their distance would be maximal. (For such applications, you probably don’t want to count stopwords such as the and in, which don’t truly signal semantic similarity.)
One way out of this conundrum is the word mover’s distance (WMD), introduced in From Word Embeddings To Document Distances, (Matt J. Kusner, Yu Sun, Nicholas I. Kolkin, Kilian Q. Weinberger, ICML 2015). WMD adapts the earth mover’s distance to the space of documents: the distance between two texts is given by the total amount of “mass” needed to move the words from one side into the other, multiplied by the distance the words need to move. So, starting from a measure of the distance between different words, we can get a principled document-level distance. Here is a visualisation of the idea, from the ICML slides:
Prepare some word embeddings¶
The key ingredient in WMD is a good distance measure between words. Dense representations of words, also known by the trendier name “word embeddings” (because “distributed word representations” didn’t stick), do the trick here. We could train the embeddings ourselves, but for meaningful results we would need tons of documents, and that might take a while. So let’s just use the ones from the word2vec team. (download link)
import os
import numpy as np
from sklearn.datasets import fetch_20newsgroups
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.cross_validation import train_test_split
if not os.path.exists("data/embed.dat"):
print("Caching word embeddings in memmapped format...")
from gensim.models.word2vec import Word2Vec
wv = Word2Vec.load_word2vec_format(
"data/GoogleNews-vectors-negative300.bin.gz",
binary=True)
fp = np.memmap("data/embed.dat", dtype=np.double, mode='w+', shape=wv.syn0norm.shape)
fp[:] = wv.syn0norm[:]
with open("data/embed.vocab", "w") as f:
for _, w in sorted((voc.index, word) for word, voc in wv.vocab.items()):
print(w, file=f)
del fp, wv
W = np.memmap("data/embed.dat", dtype=np.double, mode="r", shape=(3000000, 300))
with open("data/embed.vocab") as f:
vocab_list = map(str.strip, f.readlines())
vocab_dict = {w: k for k, w in enumerate(vocab_list)}
Reproducing the demo above¶
d1 = "Obama speaks to the media in Illinois"
d2 = "The President addresses the press in Chicago"
vect = CountVectorizer(stop_words="english").fit([d1, d2])
print("Features:", ", ".join(vect.get_feature_names()))
Features: addresses, chicago, illinois, media, obama, president, press, speaks
The two documents are completely orthogonal in terms of bag-of-words
from scipy.spatial.distance import cosine
v_1, v_2 = vect.transform([d1, d2])
v_1 = v_1.toarray().ravel()
v_2 = v_2.toarray().ravel()
print(v_1, v_2)
print("cosine(doc_1, doc_2) = {:.2f}".format(cosine(v_1, v_2)))
[0 0 1 1 1 0 0 1] [1 1 0 0 0 1 1 0] cosine(doc_1, doc_2) = 1.00
from sklearn.metrics import euclidean_distances
W_ = W[[vocab_dict[w] for w in vect.get_feature_names()]]
D_ = euclidean_distances(W_)
print("d(addresses, speaks) = {:.2f}".format(D_[0, 7]))
print("d(addresses, chicago) = {:.2f}".format(D_[0, 1]))
d(addresses, speaks) = 1.16 d(addresses, chicago) = 1.37
We will be using pyemd, a Python wrapper for Pele and Werman’s implementation of the earth mover’s distance.
from pyemd import emd
# pyemd needs double precision input
v_1 = v_1.astype(np.double)
v_2 = v_2.astype(np.double)
v_1 /= v_1.sum()
v_2 /= v_2.sum()
D_ = D_.astype(np.double)
D_ /= D_.max() # just for comparison purposes
print("d(doc_1, doc_2) = {:.2f}".format(emd(v_1, v_2, D_)))
d(doc_1, doc_2) = 0.74
Document classification¶
We will use the 20 Newsgroups classification task. Because WMD is an expensive computation, for this demo we just use a subset. To emphasize the power of the method, we use a larger test size, but train on relatively few samples.
newsgroups = fetch_20newsgroups()
docs, y = newsgroups.data, newsgroups.target
docs_train, docs_test, y_train, y_test = train_test_split(docs, y,
train_size=100,
test_size=300,
random_state=0)
Since the W embedding array is pretty huge, we might as well restrict it to just the words that actually occur in the dataset.
vect = CountVectorizer(stop_words="english").fit(docs_train + docs_test)
common = [word for word in vect.get_feature_names() if word in vocab_dict]
W_common = W[[vocab_dict[w] for w in common]]
We can then create a fixed-vocabulary vectorizer using only the words we have embeddings for.
vect = CountVectorizer(vocabulary=common, dtype=np.double)
X_train = vect.fit_transform(docs_train)
X_test = vect.transform(docs_test)
One way to proceed is to just pre-compute the pairwise distances between all documents, and use them to search for hyperparameters and evaluate the model. However, that would incur some extra computation, and WMD is expensive. Also, it’s not the most pleasant user interface. So we define some scikit-learn compatible estimators for computing the WMD.
WordMoversKNN subclasses from KNeighborsClassifier and overrides the predict function to compute the WMD between all training and test samples.
In practice, however, we often don’t know what is the best n_neighbors to use. Simply wrapping WordMoversKNN in a GridSearchCV would be rather expensive because of all the distances that would need to be recomputed for every value of n_neighbors. So we introduce WordMoversKNNCV, which, when fitted, performs cross-validation to find the best value of n_neighbors (under any given evaluation metric), while only computing the WMD once per fold, and only across folds (saving n_folds * fold_size ** 2 evaluations).
"""%%file word_movers_knn.py"""
# Authors: Vlad Niculae, Matt Kusner
# License: Simplified BSD
import numpy as np
from sklearn.metrics import euclidean_distances
from sklearn.externals.joblib import Parallel, delayed
from sklearn.neighbors import KNeighborsClassifier
from sklearn.utils import check_array
from sklearn.cross_validation import check_cv
from sklearn.metrics.scorer import check_scoring
from sklearn.preprocessing import normalize
from pyemd import emd
class WordMoversKNN(KNeighborsClassifier):
"""K nearest neighbors classifier using the Word Mover's Distance.
Parameters
----------
W_embed : array, shape: (vocab_size, embed_size)
Precomputed word embeddings between vocabulary items.
Row indices should correspond to the columns in the bag-of-words input.
n_neighbors : int, optional (default = 5)
Number of neighbors to use by default for :meth:`k_neighbors` queries.
n_jobs : int, optional (default = 1)
The number of parallel jobs to run for Word Mover's Distance computation.
If ``-1``, then the number of jobs is set to the number of CPU cores.
verbose : int, optional
Controls the verbosity; the higher, the more messages. Defaults to 0.
References
----------
Matt J. Kusner, Yu Sun, Nicholas I. Kolkin, Kilian Q. Weinberger
From Word Embeddings To Document Distances
The International Conference on Machine Learning (ICML), 2015
http://mkusner.github.io/publications/WMD.pdf
"""
_pairwise = False
def __init__(self, W_embed, n_neighbors=1, n_jobs=1, verbose=False):
self.W_embed = W_embed
self.verbose = verbose
super(WordMoversKNN, self).__init__(n_neighbors=n_neighbors, n_jobs=n_jobs,
metric='precomputed', algorithm='brute')
def _wmd(self, i, row, X_train):
"""Compute the WMD between training sample i and given test row.
Assumes that `row` and train samples are sparse BOW vectors summing to 1.
"""
union_idx = np.union1d(X_train[i].indices, row.indices)
W_minimal = self.W_embed[union_idx]
W_dist = euclidean_distances(W_minimal)
bow_i = X_train[i, union_idx].A.ravel()
bow_j = row[:, union_idx].A.ravel()
return emd(bow_i, bow_j, W_dist)
def _wmd_row(self, row, X_train):
"""Wrapper to compute the WMD of a row with all training samples.
Assumes that `row` and train samples are sparse BOW vectors summing to 1.
Useful for parallelization.
"""
n_samples_train = X_train.shape[0]
return [self._wmd(i, row, X_train) for i in range(n_samples_train)]
def _pairwise_wmd(self, X_test, X_train=None):
"""Computes the word mover's distance between all train and test points.
Parallelized over rows of X_test.
Assumes that train and test samples are sparse BOW vectors summing to 1.
Parameters
----------
X_test: scipy.sparse matrix, shape: (n_test_samples, vocab_size)
Test samples.
X_train: scipy.sparse matrix, shape: (n_train_samples, vocab_size)
Training samples. If `None`, uses the samples the estimator was fit with.
Returns
-------
dist : array, shape: (n_test_samples, n_train_samples)
Distances between all test samples and all train samples.
"""
n_samples_test = X_test.shape[0]
if X_train is None:
X_train = self._fit_X
dist = Parallel(n_jobs=self.n_jobs, verbose=self.verbose)(
delayed(self._wmd_row)(test_sample, X_train)
for test_sample in X_test)
return np.array(dist)
def fit(self, X, y):
"""Fit the model using X as training data and y as target values
Parameters
----------
X : scipy sparse matrix, shape: (n_samples, n_features)
Training data.
y : {array-like, sparse matrix}
Target values of shape = [n_samples] or [n_samples, n_outputs]
"""
X = check_array(X, accept_sparse='csr', copy=True)
X = normalize(X, norm='l1', copy=False)
return super(WordMoversKNN, self).fit(X, y)
def predict(self, X):
"""Predict the class labels for the provided data
Parameters
----------
X : scipy.sparse matrix, shape (n_test_samples, vocab_size)
Test samples.
Returns
-------
y : array of shape [n_samples]
Class labels for each data sample.
"""
X = check_array(X, accept_sparse='csr', copy=True)
X = normalize(X, norm='l1', copy=False)
dist = self._pairwise_wmd(X)
return super(WordMoversKNN, self).predict(dist)
class WordMoversKNNCV(WordMoversKNN):
"""Cross-validated KNN classifier using the Word Mover's Distance.
Parameters
----------
W_embed : array, shape: (vocab_size, embed_size)
Precomputed word embeddings between vocabulary items.
Row indices should correspond to the columns in the bag-of-words input.
n_neighbors_try : sequence, optional
List of ``n_neighbors`` values to try.
If None, tries 1-5 neighbors.
scoring : string, callable or None, optional, default: None
A string (see model evaluation documentation) or
a scorer callable object / function with signature
``scorer(estimator, X, y)``.
cv : int, cross-validation generator or an iterable, optional
Determines the cross-validation splitting strategy.
Possible inputs for cv are:
- None, to use the default 3-fold cross-validation,
- integer, to specify the number of folds.
- An object to be used as a cross-validation generator.
- An iterable yielding train/test splits.
For integer/None inputs, StratifiedKFold is used.
n_jobs : int, optional (default = 1)
The number of parallel jobs to run for Word Mover's Distance computation.
If ``-1``, then the number of jobs is set to the number of CPU cores.
verbose : int, optional
Controls the verbosity; the higher, the more messages. Defaults to 0.
Attributes
----------
cv_scores_ : array, shape (n_folds, len(n_neighbors_try))
Test set scores for each fold.
n_neighbors_ : int,
The best `n_neighbors` value found.
References
----------
Matt J. Kusner, Yu Sun, Nicholas I. Kolkin, Kilian Q. Weinberger
From Word Embeddings To Document Distances
The International Conference on Machine Learning (ICML), 2015
http://mkusner.github.io/publications/WMD.pdf
"""
def __init__(self, W_embed, n_neighbors_try=None, scoring=None, cv=3,
n_jobs=1, verbose=False):
self.cv = cv
self.n_neighbors_try = n_neighbors_try
self.scoring = scoring
super(WordMoversKNNCV, self).__init__(W_embed,
n_neighbors=None,
n_jobs=n_jobs,
verbose=verbose)
def fit(self, X, y):
"""Fit KNN model by choosing the best `n_neighbors`.
Parameters
-----------
X : scipy.sparse matrix, (n_samples, vocab_size)
Data
y : ndarray, shape (n_samples,) or (n_samples, n_targets)
Target
"""
if self.n_neighbors_try is None:
n_neighbors_try = range(1, 6)
else:
n_neighbors_try = self.n_neighbors_try
X = check_array(X, accept_sparse='csr', copy=True)
X = normalize(X, norm='l1', copy=False)
cv = check_cv(self.cv, X, y)
knn = KNeighborsClassifier(metric='precomputed', algorithm='brute')
scorer = check_scoring(knn, scoring=self.scoring)
scores = []
for train_ix, test_ix in cv:
dist = self._pairwise_wmd(X[test_ix], X[train_ix])
knn.fit(X[train_ix], y[train_ix])
scores.append([
scorer(knn.set_params(n_neighbors=k), dist, y[test_ix])
for k in n_neighbors_try
])
scores = np.array(scores)
self.cv_scores_ = scores
best_k_ix = np.argmax(np.mean(scores, axis=0))
best_k = n_neighbors_try[best_k_ix]
self.n_neighbors = self.n_neighbors_ = best_k
return super(WordMoversKNNCV, self).fit(X, y)
Overwriting word_movers_knn.py
knn_cv = WordMoversKNNCV(cv=3,
n_neighbors_try=range(1, 20),
W_embed=W_common, verbose=5, n_jobs=3)
knn_cv.fit(X_train, y_train)
[Parallel(n_jobs=3)]: Done 12 tasks | elapsed: 30.8s [Parallel(n_jobs=3)]: Done 34 out of 34 | elapsed: 2.0min finished [Parallel(n_jobs=3)]: Done 12 tasks | elapsed: 25.7s [Parallel(n_jobs=3)]: Done 33 out of 33 | elapsed: 2.9min finished [Parallel(n_jobs=3)]: Done 12 tasks | elapsed: 53.3s [Parallel(n_jobs=3)]: Done 33 out of 33 | elapsed: 2.0min finished
WordMoversKNNCV(W_embed=memmap([[ 0.04283, -0.01124, ..., -0.05679, -0.00763],
[ 0.02884, -0.05923, ..., -0.04744, 0.06698],
...,
[ 0.08428, -0.15534, ..., -0.01413, 0.04561],
[-0.02052, 0.08666, ..., 0.03659, 0.10445]]),
cv=3, n_jobs=3, n_neighbors_try=range(1, 20), scoring=None,
verbose=5)
print("CV score: {:.2f}".format(knn_cv.cv_scores_.mean(axis=0).max()))
CV score: 0.38
print("Test score: {:.2f}".format(knn_cv.score(X_test, y_test)))
[Parallel(n_jobs=3)]: Done 12 tasks | elapsed: 32.2s [Parallel(n_jobs=3)]: Done 66 tasks | elapsed: 4.3min [Parallel(n_jobs=3)]: Done 156 tasks | elapsed: 12.5min [Parallel(n_jobs=3)]: Done 282 tasks | elapsed: 30.5min [Parallel(n_jobs=3)]: Done 300 out of 300 | elapsed: 48.9min finished
Test score: 0.31
Comparison with other models¶
Now let’s see how WMD compares with some common approaches, on bag of words features. The most apples-to-apples comparison would be K nearest neighbors with a cosine similarity metric. This approach performs worse than using WMD. (All scores are accuracies.)
from sklearn.svm import LinearSVC
from sklearn.neighbors import KNeighborsClassifier
from sklearn.grid_search import GridSearchCV
knn_grid = GridSearchCV(KNeighborsClassifier(metric='cosine', algorithm='brute'),
dict(n_neighbors=list(range(1, 20))),
cv=3)
knn_grid.fit(X_train, y_train)
print("CV score: {:.2f}".format(knn_grid.best_score_))
print("Test score: {:.2f}".format(knn_grid.score(X_test, y_test)))
CV score: 0.34 Test score: 0.22
Another common method for text classification is the linear support vector machine on bag of words. This performs a bit better than vanilla cosine KNN, but worse than using WMD in this setting. In our experience, this seems to depend on the amount of training data available.
svc_grid = GridSearchCV(LinearSVC(),
dict(C=np.logspace(-6, 6, 13, base=2)),
cv=3)
svc_grid.fit(X_train, y_train)
print("CV score: {:.2f}".format(svc_grid.best_score_))
print("Test score: {:.2f}".format(svc_grid.score(X_test, y_test)))
CV score: 0.35 Test score: 0.27
What have we learned?¶
WMD is much better at capturing semantic similarity between documents than cosine, due to its ability to generalize to unseen words. The SVM does somewhat better than cosine KNN, but still lacks such out-of-vocabulary generalization. Given enough data, WMD can probably improve this margin, especially using something like metric learning on top.
The exact WMD, as we have used it here, is pretty slow. This code is not optimized as much as it could be, there is potential through caching and using Cython. However, a major limitation remains the cost of actually computing the EMD. To scale even higher, exactness can be relaxed by using lower bounds. In our next post, we will compare such optimization strategies, as discussed in the WMD paper.
Comments !